IAP-25-034
Reproductive isolation in a species group of Drosophila
The fruit fly Drosophila melanogaster has been used as a model organism for over 100 years, but increasingly evolutionary biologists are examining natural variation within and between different species of the fruit fly to understand adaptation and speciation. The virilis group of Drosophila contains twelve species which differ in behaviour, chromosome structure and genomic features, and occupy a wide range of different global ecologies, including extreme cold adaptation. The studentship will use recently obtained and new genomic data, combined with laboratory crosses and experiments, to examine the evolution of reproductive isolation across the whole group for the first time since a classic study by Throckmorton in 1982. Combined with the new genomic data the project can address important novel questions such as the rate of evolution of pre- versus post-mating isolation and how these are correlated with genomic features such as the extent of chromosomal inversion variation between species, variation in the structure of the X chromosome, and unusual patterns in the distribution of transposable elements. Also we can ask if ecological specialisation leads to different types of reproductive isolation; for example, divergence in cuticular hydrocarbons can occur with climate adaptation but these also act as contact pheromones. This is a very “open” studentship with much potential for the student to develop the project along their own interests.
Click on an image to expand
Image Captions
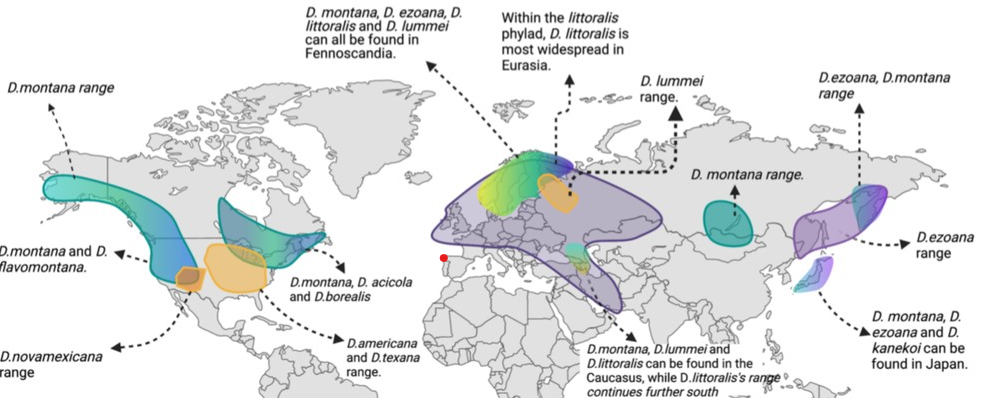
Biogeography of the virilis group
Methodology
The student will receive broad training in Evolutionary Biology, genomics, speciation biology and bioinformatics. Their will be training in handling and analysing genome scale data to extract a range of data from existing genomes, and hopefully augmenting these with new genomes, both short and long read sequencing to allow examining of genomic structure (e.g. the presence of inversions and characterisation of the sex chromosome. The student will also complete laboratory studies of Drosophila, including crosses and the scoring of behavioural data, including assortative mating analysis and the characterisation of mating characteristics such as pheromones and male courtship songs.
Project Timeline
Year 1
Basic bioinformatic analysis of genomes, preparation of new genomes and preliminary crosses of all species.
Year 2
Detailed characterisation of pre- and postmating isolation across species and correlation analysis with genomic features discussed in project description.
Year 3
Characterisation of ecological variation across species, potential analyses of ecological adaptation (hot and cold-tolerance). Potential analyses of gene flow between contemporary wild populations.
Year 3.5
Detailed statistical analyses and paper writing.
Training
& Skills
The student will receive advanced training in evolutionary biology, genomics, speciation and bioinformatics.
References & further reading
Yusuf, L., et al. 2022. Divergence and introgression among the virilis group of Drosophila. Evolution Letters 6: 537–551. https://doi.org/10.1002/evl3.301
Poikela, N., et al. 2024. Chromosomal inversions and the demography of speciation in Drosophila montana and Drosophila flavomontana. Genome Biology and Evolution 16(3). https://doi.org/10.1093/gbe/evae024
Flynn, J. M., et al. 2024. High-Quality genome assemblies reveal evolutionary dynamics of repetitive DNA and structural rearrangements in the Drosophila virilis subgroup.” Genome Biology and Evolution 16. https://doi.org/10.1093/gbe/evad238
Throckmorton, L. H. 1982. The virilis species group. Genetics and Biology of Drosophila 3b: 227-296.
Shaw, et al. 2024. How Important Is Sexual Isolation to Speciation? Cold Spring Harbor Perspectives in Biology 16 DOI 10.1101/cshperspect.a041427

